The crystalline structure of F 1,6-BPase of thermophillic archaea, Sulfolobus
tokodaii was obtained by X-ray diffraction (figures 2 & 3) that show
the alpha and beta sheets. F 1,6-BPase is a 385 amino acid long hypothetical
protein with formula mass of 40122.6 Daltons. It has a residue conservation
of 359 identified residues on chain A. The structure of F 1,6-BPase is
made up of 3 beta sheets, 6 beta hairpins, 7 beta bulges, 15 strands, 14
helices, 6 helix-helix interactions, 41 beta turns, and 4 gamma turns (4).
F 1,6-BPase is a tetrameric enzyme of four identical subunits each of 337
amino acid residues and molecular weight of about 36,500 Daltons. On each
subunit is an active site for the substrate (2FP), 28 Angstroms away from
the active site is an AMP binding site, and a metal binding site lies between
these two domains (5). The four subunits of F 1,6-BPase are labeled from
upper left to right (clockwise) C1, C2, C3, and C4 respectively. C1 and
C2 correspond to the upper dimer while C3 and C4 correspond to the lower
dimer. The tertiary structure of each subunit is divided into two domains,
the AMP domain (residues 1–200) and the F 1,6-bisphosphate domain (residues
201–337). The AMP domain has the AMP binding site at the C1–C4 interface
and the Fructose 1,6 bisphosphate domain contains the active site at the
C1–C2 interface (6). |
 |
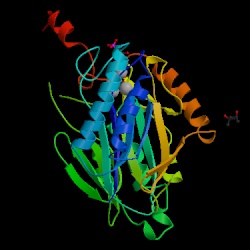 |
| Figure 2. Crystal structure of
the fructose 1,6-bisphosphatase (Biological molecule unit) of Sulfolobus
tokodaii. Nishimasu H. et al. RCSB
Protein Data Bank. |
Figure 3. Crystal structure of
the fructose 1,6-bisphosphatase (Asymmetrical unit) of Sulfolobus tokodaii.
Nishimasu H. et al. RCSB
Protein Data Bank |
|